On February 6 (Beijing Time), a research team led by Professor Zhang Jisen from the School of Agriculture at Guangxi University(GXU), the Guangxi Laboratory for Sugarcane Biological Breeding, and the State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources published a research article in Science, titled “Multiscale pangenome graphs empower the genomic dissection of mixed-ploidy sugarcane species.” The study was conducted in collaboration with Fujian Agriculture and Forestry University and other institutions.
GXU is the first corresponding institution and one of the co-first-author affiliations of the paper. The co-first authors, all members are in Professor Zhang Jisen’s team, they are Huang Yumin (Ph.D., Fujian Agriculture and Forestry University), Zhang Yixing (Research Assistant, GXU), and Professor Zhang Qing (GXU). Professor Zhang Jisen is the first corresponding author, and Professor Tang Haibao of Fujian Agriculture and Forestry University serves as co-corresponding author.
This study is the first to propose and implement a polyploid-aware, multiscale (genome–gene–protein) graph pangenome analytical framework. The approach provides a critical technological breakthrough for the genetic dissection and molecular breeding of complex polyploid crops, carrying significant scientific importance and substantial potential for industrial application. It marks a major advancement by Chinese researchers in the field of sugarcane graph pangenomics and polyploid genetic analysis.
This achievement represents another major milestone in Professor Zhang Jisen’s long-term research program centered on sugarcane biological breeding. Focusing on a systematic research chain spanning sugarcane genomics, germplasm resource evolution, polyploid genetics, and the identification of genes with significant breeding value, the team has previously published a series of landmark studies in Nature Genetics (2018, 2022, 2025) and Nature Plants (2023). The present work further consolidates their leading position in this field.
Sugarcane (Saccharum spp.) is a globally important sugar and bioenergy crop, accounting for approximately 80% of the world’s sugar production and 40% of ethanol output. It is widely regarded as one of the most promising renewable energy crops. Guangxi is the largest sugarcane-producing region in China. With domestic sugar demand continuing to rise, China’s sugar supply gap is projected to reach approximately 13 million tons by 2030. The sustainable development of the sugarcane industry is therefore of strategic importance for ensuring national sugar security.
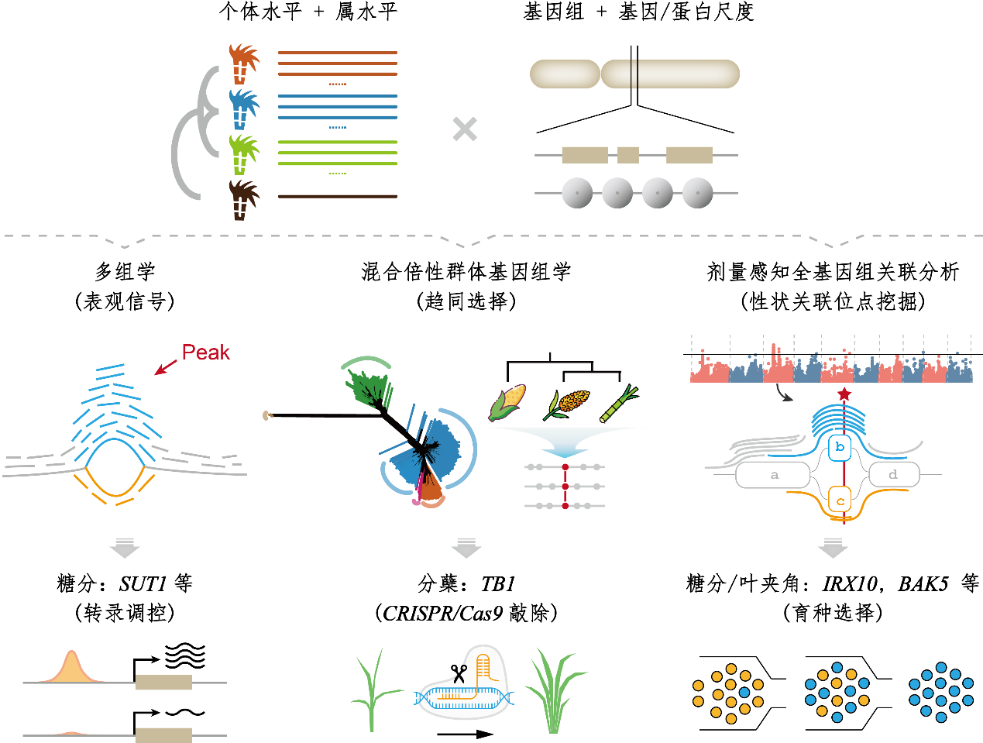
Figure. Graph pangenome strategy for complex polyploid sugarcane enhances the discovery of key breeding genes
Modern cultivated sugarcane varieties originated from interspecific hybridization and backcrossing between high-sugar tropical Saccharum officinarum and stress-tolerant wild Saccharum spontaneum more than a century ago. As a result, the sugarcane genome exhibits extreme complexity, characterized by high ploidy levels, large chromosome numbers, frequent structural variations, and a high abundance of repetitive sequences. Such features make it difficult for a traditional single linear reference genome to accurately resolve the genetic architecture and allelic dosage of sugarcane. In practice, sequencing data often suffer from alignment challenges and information loss during analysis. Increasing evidence suggests that the conventional “single linear genome” paradigm is insufficient for studying complex polyploid crops like sugarcane, highlighting the urgent need for new reference systems and analytical frameworks.
To address this challenge, building upon their previous successful assembly of an ultra-complex genome of modern cultivated sugarcane and a gap-free, high-quality genome of the related species Erianthus fulvus, the research team moved beyond the traditional single-reference paradigm and established a polyploid-aware graph pangenome framework. By integrating nine genome assemblies from four sugarcane species, the graph captured approximately 82% of the genetic diversity—substantially higher than the about 34% coverage achieved by a single reference genome. Importantly, the advantages of this strategy extend beyond improved data coverage; it establishes a unified and unbiased analytical framework for genomic studies across populations and ploidy levels.
The team further developed DosageGWAS, a novel strategy for dissecting dosage effects in polyploid genomes. Compared with conventional linear GWAS approaches, DosageGWAS demonstrated clear advantages in heritability estimation and trait-locus detection power. Using this method, the researchers successfully identified multiple candidate genes closely associated with sugar content and leaf angle traits, and systematically elucidated the genetic basis of key agronomic characteristics, including sugar metabolism.
Cross-species comparative analysis within the Sorghum clade further revealed convergent selection between sugarcane and sorghum in pathways related to carbohydrate metabolism and hormone response. Functional validation using CRISPR technology confirmed that TB1 is a key regulator of tillering in sugarcane. High-efficiency knockout lines exhibited significantly increased tiller numbers, earlier tiller initiation, and enhanced yield performance. Together, these findings establish a complete research pipeline from “graph-based discovery” to “functional validation” and ultimately to the identification of actionable breeding targets.
Overall, the study overcomes major technical bottlenecks in polyploid genetic analysis, precisely identifies genes and regulatory elements associated with important agronomic traits such as tillering and sugar accumulation, and directly supports molecular breeding for high-sugar and high-yield sugarcane varieties. Association analyses of sugar-related traits were conducted across more than one hundred germplasm accessions, achieving substantial progress in capturing genetic diversity, mapping key traits, and mining breeding targets. The framework provides core technological support for molecular design breeding in complex polyploid crops and holds broad application prospects.
This work was jointly supported by the National Key Research and Development Program of China, the Guangxi Major Science and Technology Program, the Guangxi Key R&D Program, and the National Natural Science Foundation of China.